Clades of interest to be plotted in color
Arguments
- Tibble
A
ConvenientTblTreeobject.- left, right
Leaf labels whose most common recent ancestor will define the clade
- color
Color of the clade in plots
- displayName
Display name of the clade (can contain spaces)
See also
Other Focal clade functions:
eog(),
recordAncestor(),
recordClades(),
subMatrix(),
subTree(),
visualizeTree()
Examples
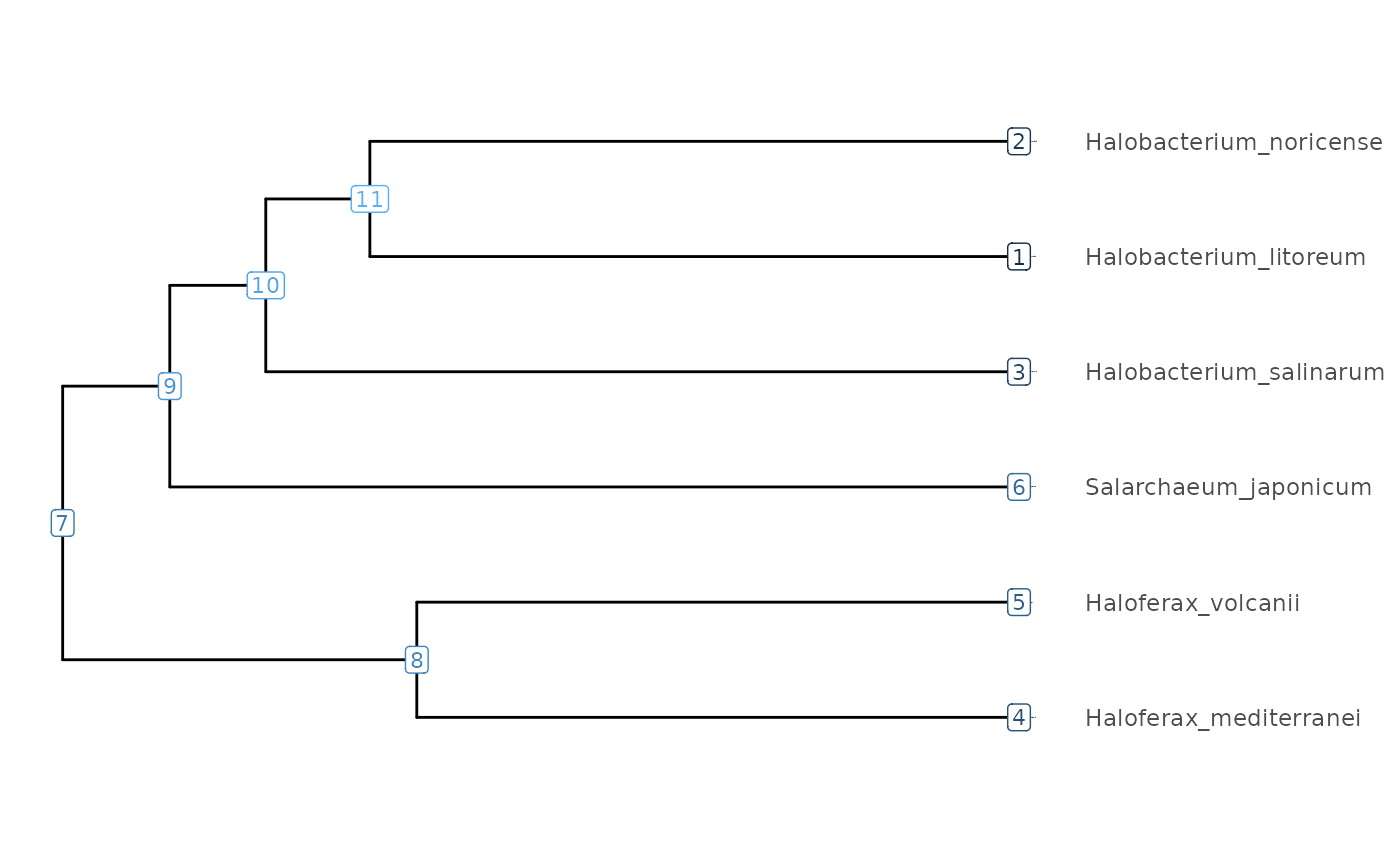
(Halobacterium <- focalClade(Halo_Tree,
"Halobacterium_noricense", "Halobacterium_salinarum", "blue", "Halobacterium genus"))
#> Halobacterium genus, node ID: 10, number of genomes: 3
(Haloferax <- focalClade(Halo_Tree,
"Haloferax_mediterranei", "Haloferax_volcanii", "green3", "Haloferax genus"))
#> Haloferax genus, node ID: 8, number of genomes: 2
(clades <- FocalCladeList(Halobacterium, Haloferax))
#> Halobacterium genus, node ID: 10, number of genomes: 3
#> Haloferax genus, node ID: 8, number of genomes: 2
visualizeTree(Halo_Tree) + clades