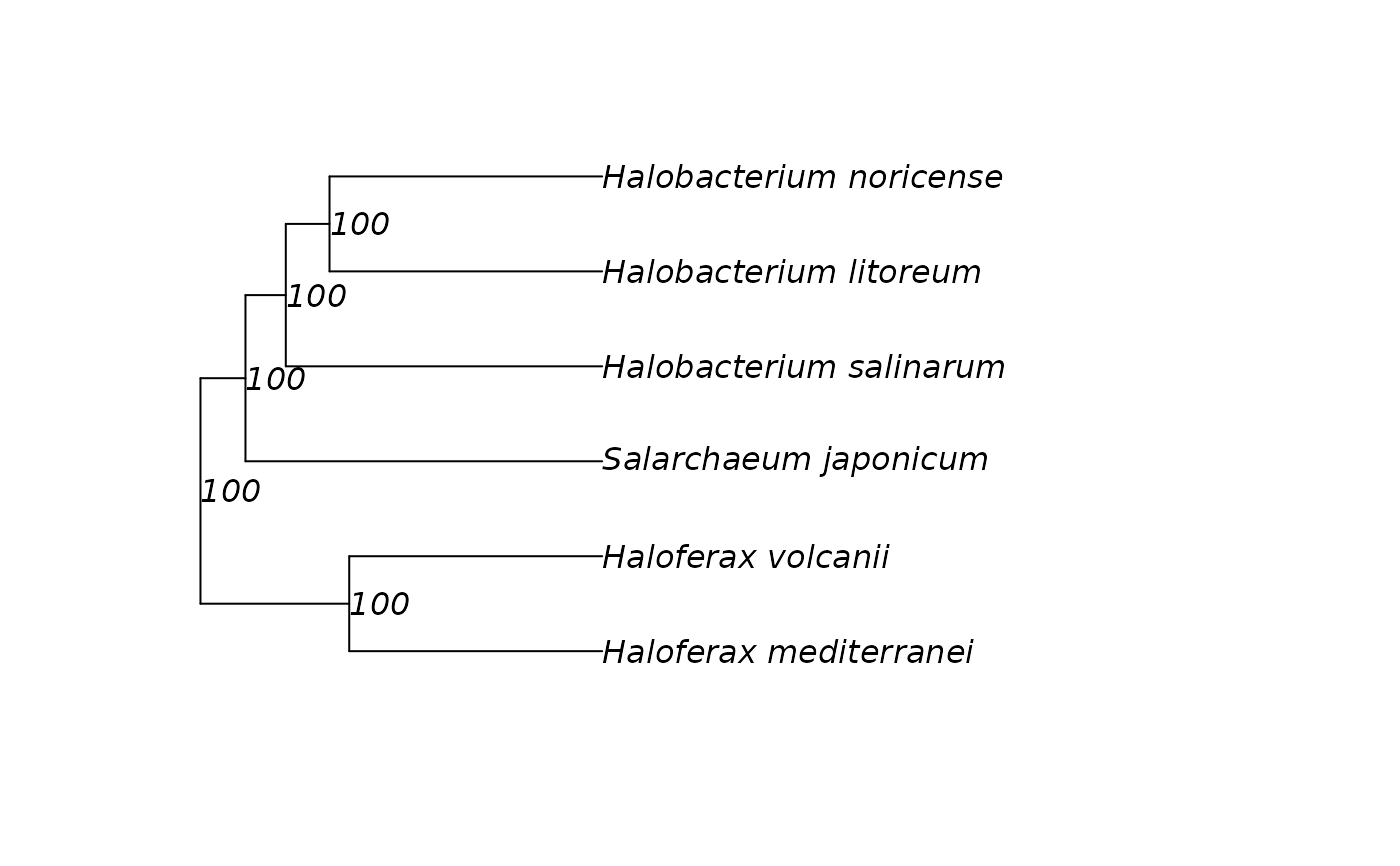
Residual-resampling bootstrap tree(s) from a distance matrix (UPGMA)
Source:R/bootstrapTree.R
residualBootstrapTree.RdBuild a reference tree from a distance matrix using UPGMA, then perform residual-resampling bootstrap to obtain replicate trees and map clade support onto the reference.
Value
A list with:
tree: Reference tree (phylo) built fromDwith UPGMA. Node labels contain bootstrap %.bootstrap_trees: List of replicate trees (phylo).support: Numeric vector of bootstrap percentages per internal node (aligned totree$node.label).call: Matched call.
Examples
out <- residualBootstrapTree(Halo_PercentDiff, n_bootstrap = 200)
plot(out$tree, show.node.label = TRUE)